GET IN TOUCH
Have questions?
Contact our client services team today.
- services@mplnet.com
- 865.380.9746
- 250 E. Broadway Maryville, TN 37804
Molecular Pathology Laboratory Network, Inc. (MPLN) has been at the forefront of laboratory medicine since its inception. Furthermore, our molecular oncology diagnostics (MDX) laboratory performs quantitative polymerase chain reaction (qPCR) and Next Generation Sequencing (NGS) analyses to support therapy selection, treatment, monitoring, and minimal residual disease detection.

NGS enables the identification of genetic sequences within an individual’s whole genome. This powerful technique is widely used in molecular oncology for classifying key gene sequences and hotspot mutations, providing context for:
IDENTIFYING:
ENABLING:
NGS can greatly benefit patients with new diagnoses or resistance to conventional therapy, as well as those facing complex diagnostic challenges. Furthermore, as the range of genetic mutations associated with cancer that can be targeted with treatments continues to expand, MPLN is committed to offering patients and clients the latest laboratory-developed tests (LDTs) and FDA-approved in vitro diagnostic (IVD) assays.
NGS offers significant advantages over PCR based approaches for the detection and monitoring of lymphoproliferative T-cell and B-cell disorders. Offerings include:
Although quantitative PCR (qPCR) based methodology is more than 20 years old, it remains a widely used inexpensive methodology for the detection of individual genetic mutations with sensitivity as low as 0.001%. At MPLN, both laboratory developed tests (LDTs) and FDA approved (IVD) assays are used for a wide variety of applications including:
Advances in pharmacogenetics, along with the design and
development of targeted cancer therapies, have led to an
increasing number of companion diagnostic assays.
Whereas pharmacogenetics explores a patient’s specific
genotype and response to a drug, companion diagnostics is a
broader application that includes the association of biomarkers
and genetic mutations that predict treatment outcomes.
Using molecular technology, we can identify patients who may
experience adverse drug interactions or those who may be
unresponsive to certain medications due to genetic variations or
expression of specific hormones, proteins and/or enzymes.
Identifying gene mutations and specific markers prior to
initiating therapy can result in better patient outcomes as well
as substantial cost savings by avoiding poor drug choices and
needless over prescribing.
For example, colorectal cancer (CRC) patients with KRAS or
BRAF mutations are less likely to respond to anti-EGFR therapy.
Therefore, ASCO recommends patients with metastatic CRC who
are candidates for anti-EFGR therapy be tested for KRAS gene
mutations prior to initiating treatment. Also, when the KRAS
gene is not mutated, NCCN guidelines recommend determination
of BRAF gene status as part of their workup, although this
recommendation is currently based on inconsistent field data.
T-cell receptor and B-cell immunoglobulin heavy chain (IgH) or
light chain (Igk) gene rearrangement studies can be supportive
of a lymphoma diagnosis with identification of monoclonality in a
morphologically suspicious lymphoid infiltrate.
Molecular immunoglobulin kappa (Igk) light chain testing is
also a useful complement to B-cell heavy chain (IgH) gene
rearrangement analysis. Igk gene rearrangement can provide confirmation of clonality in post-germinal center (mature)
neoplasms where clonotypic IgH signatures may not be detected
due to somatic hypermutation of VH genes.
The Igk assay, in addition to identifying clonality in atypical
lymphoproliferative disorders, can also support a differential
diagnosis between reactive lesions and hematologic
malignancies, and assign presumptive lineage in mature
monoclonal lymphoproliferative disorders.
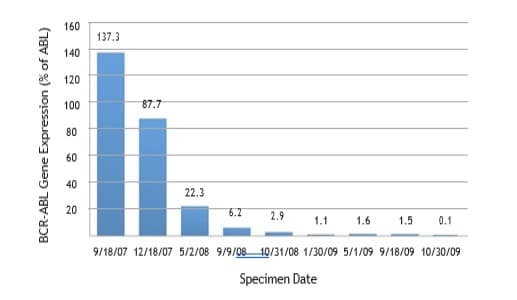
Detection and monitoring of minimal residual disease can
be accomplished by quantitative PCR, specifically in chronic
myelogenous leukemia and acute prolymphocytic leukemia.
Quantitative PCR provides at least 100 to 1000 times greater
sensitivity than fluorescent in situ hybridization (FISH).
Measurements are taken at baseline in newly diagnosed cases and
at regular intervals to monitor for evidence of molecular remission in
response to chemotherapy or allogeneic stem cell transplantation.
Studies using gene sequencing can also assist in identifying patients
with an acquired drug resistance to tyrosine kinase inhibitor (TKI)
therapy, and NCCN guidelines recommend consideration of ABL
Kinase domain sequencing for these patients.
| Molecular Oncology Test | Diagnostic Condition & Companion Therapy |
| ABL kinase mutation analysis | Philadelphia positive leukemia (CML, ALL, AML) and tyrosine kinase inhibitor resistance |
| AML mutation profile: FLT3 and NPM1 mutations with reflex to CEBPA mutation | AML |
| BCR/ABL quantitative PCR major and minor | Philadelphia positive leukemia (CML, ALL, AML), baseline, and monitoring minimal residual disease |
| B-cell heavy chain gene rearrangement B-cell kappa light chain gene rearrangement |
B-cell clonality |
| BRAF V600E mutation | EGFR inhibitor response in metastatic Colorectal Cancer (CRC): Erbitux® (cetuximab), and Vectibix™ (panitumumab) |
| c-kit mutation | AML |
| JAK2 V617F mutation | PV |
| KRAS mutation | EGFR inhibitor response in metastatic CRC: Erbitux, and Vectibix |
| MPL W515/S505N mutation | EV, PMF |
| PML/RARA t(15;17) quantitative PCR short and long form | APL, baseline and monitoring minimal residual disease |
| T-cell gamma receptor gene rearrangement | T-cell clonality |
| UGT1A1 genotype | Camptosar® (Irinotecan) toxicity and drug dosage in metastatic or recurrent CRC |
| *Fluorescence in situ hybridization (FISH) assays | *Complement molecular PCR assays for diagnosis, prognosis and minimal residual disease detection |
*Fluorescence in situ hybridization (FISH) assays performed in our
Cytogenetics Laboratory complement molecular PCR assays for
diagnosis, prognosis and minimal residual disease detection of
hematological malignancies and solid tumors. Clinical indications
and/or results from previous testing are utilized to select specific
gene locations for investigation. FISH uses fluorescent probes that
bind to only those parts of the chromosome with which they show a
high degree of sequence similarity.
For a complete list of FISH probes available to detect and localize the
presence or absence of specific DNA sequences on
chromosomes in metaphase, interphase cells or in tissue, visit us
online at www.MPLNET.com.
References
1. Ross J S et al. (2004). Targeted Therapies for Cancer 2004. Am J Clin Pathol.122(4):598-609.
Trademarks
Gleevec is a registered trademark of Novartis Pharmaceuticals Corporatiwon.
Erbitux is a registered trademark of Imclone systems, Inc.
Vectibix is a trademark of Amgen, Inc.
Camptosar is a registered trademark of Pfizer, Inc
Contact our client services team today.





Our advanced technology enables simultaneous visualization of single or multiple biomarkers within the same tissue section, delivering comprehensive insights and precise diagnostics. With unmatched expertise and cutting-edge technology, our team supports you in understanding complex tissue microenvironments, aiding patient stratification and treatment response prediction.



